Clostridium Knock-Out
Knock-Out Vectors
Knock-out vectors are available for the generation of mutants in a classic two-step process by allelic exchange [1], where either replication deficient (suicide) or defective (pseudo-suicide) vectors carrying appropriate mutant alleles integrate into the genome via homologous recombination, and the double crossover mutants arise by plasmid excision in which the wildtype allele has exchanged with the mutant allele formerly on the plasmid – allelic exchange.
The isolation from single crossover integrants of those cells that have lost the plasmid to form double crossover mutants is reliant on the use of the counter selection markers codA or pyrE [1]. They may only be used in cells that lack functional equivalents, either naturally occurring or deliberately created. These are available from the SBRC online shop.
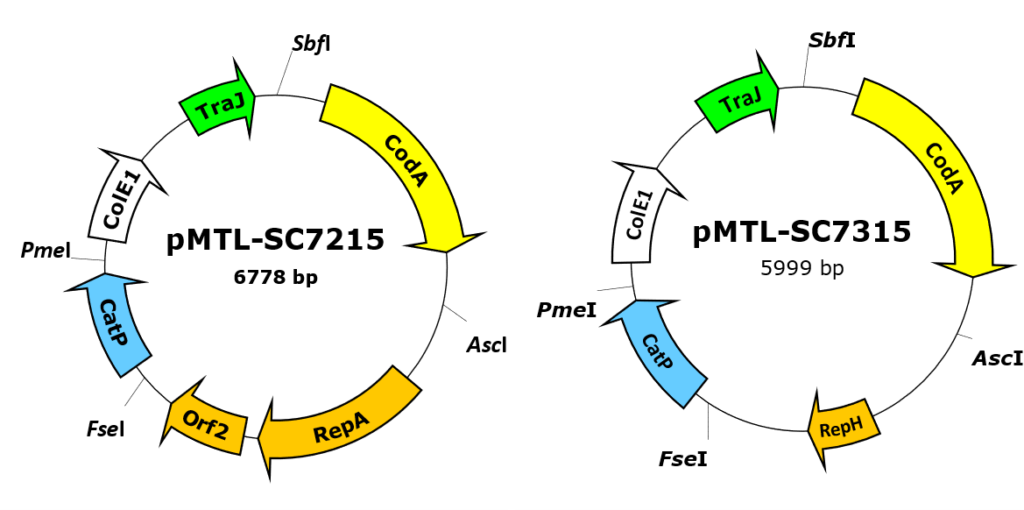
[1] codA-based plasmids (pMTL-SC7215, pMTL-SC7315 & pMTL-SC7515)
Based on the counter selection marker codA encoding cytosine deaminase which catalyzes the conversion of 5-fluorocytosine (FC) into the highly toxic 5-fluorouracil (FU). Plasmid SC7215 relies on the pBP1 replicon for the most effective pseudo-suicide in C. difficile R20291, SC7315 incorporates the pCB102 replicon for use in C. difficile 630 [2] and SC7315 is based on the pIM13 replicon for use in C. acetobutylicum [3].
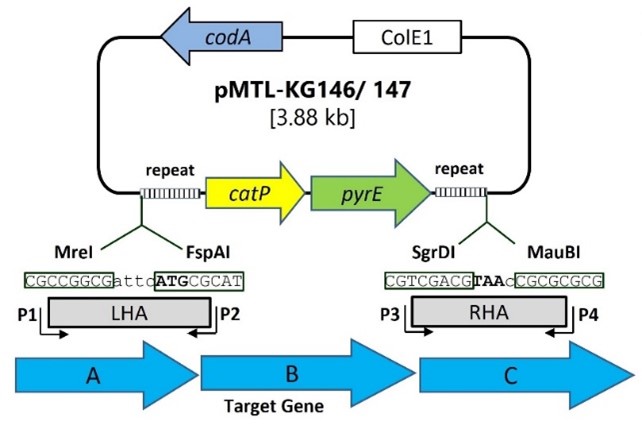
[2] codA– and pyrE-based plasmids (pMTL-KG146/147)
A suicide system for gene knock-out in C. acetobutylicum [4], relying on ColE1 for replication in E.coli and the catP for selection (chloramphenicol resistance). The counterselection marker is codA (selecting on 5-FC). The presence of pyrE and flanking repeat sequences, allows for the direct selection of clean, in-frame deletions by plating on FOA, and so the system can only be used in a pyrE deletion strain of C. acetobutylicum.
[3] pyrE-based plasmids
The pyrE gene encodes orotate phosphoribosyltransferase, an enzyme involved in de novo pyrimidine biosynthesis and which renders cells sensitive to 5-fluoro-orotatic acid (FOA). Heterologous pyrE genes are employed in specific, uracil auxotrophs created by deleting the 3’-end of the native pyrE gene using Allele-Couple Exchange (ACE) [5].
pMTL-ME3 & pMTL-GL3
Based on the pIM13 replicon and a C. spororgenes pyrE gene, pMTL-ME3 was developed for the creation of mutants in C. acetobutylicum [3]. Plasmid pMTL-GL3 was derived from pMTL-ME3 through deletion of the pIM13 replicon and shown to function as a suicide system in C. beijerinckii [6].

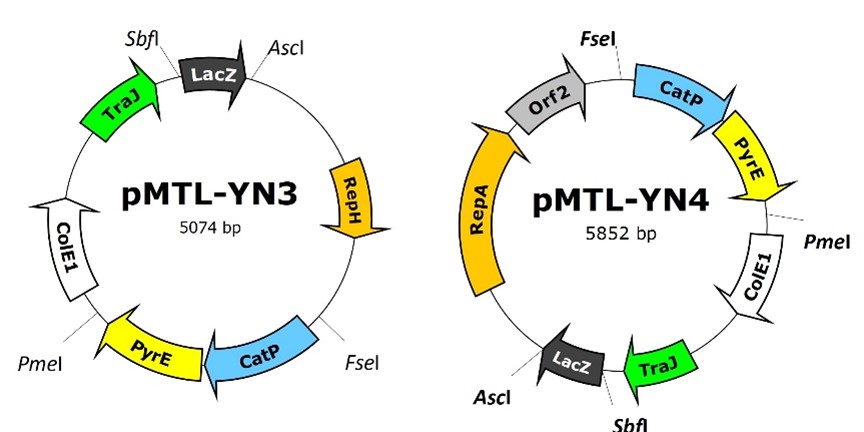
pMTL-YN3 and pMTL-YN4
Based on the pCB102 replicon (pMTL-YN3) or the pBP1 replicon (pMTL-YN4) and a C. spororgenes pyrE gene, the two KO vectors were designed for use in C. difficile R20291 and in C. difficile 630, respectively [7].
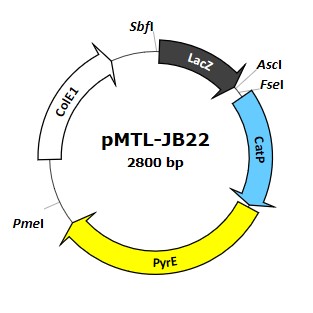
pMTL-JPB22
A suicide system for gene-knockout strain production in A. woodii [8,9,10], relying on ColE1 for replication in E.coli and the catP for selection (chloramphenicol resistance). The counterselection marker is pyrE (selecting on FOA), and so the mutant can only be used in a pyrE deletion strain of A. woodi.
References
[1] Minton NP, Ehsaan M, Humphreys CM, Little GT, Baker J, Henstra AM, Liew F, Kelly ML, Sheng L, Schwarz K, Zhang Y. A roadmap for gene system development in Clostridium. Anaerobe. 2016;41:104-112.
https://doi.org/10.1016/j.anaerobe.2016.05.011
[2] Cartman ST, Kelly ML, Heeg D, Heap JT, Minton NP. Precise manipulation of the Clostridium difficile chromosome reveals a lack of association between the tcdC genotype and toxin production. Appl Environ Microbiol. 2012;78:4683-90.
https://doi.org/10.1128/AEM.00249-12
[3] Ehsaan M, Kuit W, Zhang Y, Cartman ST, Heap JT, Winzer K, Minton NP. Mutant generation by allelic exchange and genome resequencing of the biobutanol organism Clostridium acetobutylicum ATCC 824. Biotechnol Biofuels. 2016;9:4.
https://doi.org/10.1186/s13068-015-0410-0
[4] Omorotionmwan BB, Wang H, Baker JP, Gizynski K, Yoo M, Akaluka C, Zhang Y, Minton NP. Chromosomal engineering of inducible isopropanol- butanol-ethanol production in Clostridium acetobutylicum. Front Bioeng Biotechnol. 2023;11:1218099.
https://doi.org/10.3389/fbioe.2023.1218099
[5] Heap JT, Ehsaan M, Cooksley CM, Ng YK, Cartman ST, Winzer K, Minton NP. Integration of DNA into bacterial chromosomes from plasmids without a counter-selection marker. Nucleic Acids Res. 2012;40(8):e59.
https://doi.org/10.1093/nar/gkr1321
[6] Little GT, Willson BJ, Heap JT, Winzer K, Minton NP. The Butanol Producing Microbe Clostridium beijerinckii NCIMB 14988 Manipulated Using Forward and Reverse Genetic Tools. Biotechnol J. 2018;13(11):e1700711.
https://doi.org/10.1002/biot.201700711
[7] Ng YK, Ehsaan M, Philip S, Collery MM, Janoir C, Collignon A, Cartman ST, Minton NP. Expanding the repertoire of gene tools for precise manipulation of the Clostridium difficile genome: allelic exchange using pyrE alleles. PLoS One. 2013;8(2):e56051.
https://doi.org/10.1371/journal.pone.0056051
[8] Westphal L, Wiechmann A, Baker J, Minton NP, Müller V. The Rnf Complex Is an Energy-Coupled Transhydrogenase Essential To Reversibly Link Cellular NADH and Ferredoxin Pools in the Acetogen Acetobacterium woodii. J Bacteriol. 2018 Oct 10;200(21):e00357-18.
https://doi.org/10.1128/JB.00357-18
[9] Schoelmerich MC, Katsyv A, Sung W, Mijic V, Wiechmann A, Kottenhahn P, Baker J, Minton NP, Müller V. Regulation of lactate metabolism in the acetogenic bacterium Acetobacterium woodii. Environ Microbiol. 2018 Dec;20(12):4587-4595.
https://doi.org/10.1111/1462-2920.14412
[10] Baker JP, Sáez-Sáez J, Jensen SI, Nielsen AT, Minton NP. A clean in-frame knockout system for gene deletion in Acetobacterium woodii. J Biotechnol. 2022; 353:9-18.
https://doi.org/10.1016/j.jbiotec.2022.05.013
